> Assays per cell type > Human Pluripotent Stem Cells (hPSCs)
Which Quality Control Assays
for Human Pluripotent Stem Cells (hPSCs)
Human Pluripotent Stem Cells (hPSC) have opened up an incredible amount of possibilities in the world of cell therapy. However, they require a robust quality control to be in place before one can reap the rewards of these amazing cells. Challenges like genomic instability, cell line cross-contamination, mycoplasma and pluripotency or differentiation efficiency need to be addressed effectively.
Stem Genomics is here to help and will accompany you all the way to ensure you gain confidence in the quality, reproducibility and robustness of your research work.
Discover a wide range of essential quality control assays for hPSCs
Detect over 93% of recurrent abnormalities in hPSCs in record-breaking time !
Ensure consistent cell line identity throughout your culture
Quickly monitor the differentiation and trilineage profile
The ultimate karyotype solution for hPSC in-process genomic stability testing
Look deeper into any human cell. Detect both SNVs and CNVs
Confidently detect mycoplasma in any human cell culture
Combine up to 5 essential tests in one-go!
Contact us for a customized assay
Not sure which test could best meet your needs? We’re here to help.
Genomic stability
Genomic stability is one of the primary concerns for research scientists working on human pluripotent stem cells. Although these cells have amazing capabilities, they are prone to developing chromosomal abnormalities during their time in culture. This can also be the case after critical bottleneck events such as gene editing, basically every time the cells experience replicative stress imposed by in-vitro culture.
The chromosomal defects you need to watch for when you are a research scientist working on hPSCs
Recurrent genomic alterations, essentially copy number variations (CNVs), often affect the same genomic regions, such as 1q, 12p, 17q, 20q and the X chromosome. This provides the affected cells with a selective advantage which may favor proliferation, cell survival or reduce cell differentiation capacities.
Gain of 20q11.21 copy-number variant (CNV) is detected in more than 20% of human Pluripotent Stem Cells (hPSCs) cultured worldwide and represents 22.9% of the recurrent structural variants identified in hPSCs (Assou et al., 2020; Avery et al., 2013; Halliwell et al., 2020). This makes it the most common genomic abnormality in hPSCs. Unfortunately, it cannot always be detected by conventional methods.
Concurrently, as cell therapy evolves and an increasing number of studies reach the clinical stage, demand increases for more exhaustive analyses of copy number variations (CNVs) across the entire genome. With the advent of cutting-edge technologies such as next-generation sequencing (NGS), the ability to detect novel abnormalities has greatly improved. Many recurrent CNV regions previously described using these less sensitive techniques are now being re-evaluated and redefined with greater accuracy using NGS-based approaches. This improved sensitivity and resolution in CNV analysis is making a significant contribution to our understanding of genomic variation and its potential impact in various clinical and research settings.
Beyond CNV detection, it is also critical to look for Single Nucleotide Variations and Indels in hPSCs to properly check your cells’ genomic stability. Genetic variants associated with cancer are of particular interest to the stem cell community since they might alter the cells’ genomic stability and growth characteristics, disrupt differentiation, or increase the risk of cancerous growths arising after transplant (Merkle et al. 2022).
In the case of human Pluripotent Stem Cells, the abnormalities that arise will persist following differentiation. Whether you differentiate your cells or not, this can invalidate your research, interfere with studies that use iPSC cells for disease modelling or drug screening or present substantial risks for patient health in clinical trials.
Finally, the other important factor to take into account when working on hPSCs is the speed at which those alterations can overcome the whole culture (Hastings et al., 2009). It can happen in 5 passages or less (Assou et al., 2020; McIntire et al., 2020; Pamies et al., 2017). It is therefore essential to perform tests routinely, and at early stages.
It is therefore crucial for researchers to undertake regular in-process controls to identify recurrent (or frequent) CNVs (such as 20q gain) and to consider sequencing to detect SNV abnormalities (such as TP53 mutations). These combined methods will help scientists secure their hPSC cultures.
A range of 3 possible genomic tests to meet your needs:
Detect over 93% of recurrent abnormalities in hPSCs in record-breaking time
The ultimate combined solution for hPSC in-process genomic stability testing
Look deeper into any human cell. Detect both SNVs and CNVs
Not sure which test could best meet your needs? We’re here to help.
Effective genomic stability testing for hPSCs implies a fast and cost-effective assay:
In order to follow recommended guidelines from the ISSCR, a suitable test needs to be selected. Although pertinent at certain key stages of the workflow, G-Banding, the technique traditionally used for genomic stability testing, is not suitable for regular in-process assessment. It would slow processes down and be cost-prohibitive. With these constraints in mind, Stem Genomics has developed a rapid and cost-effective digital PCR-based solution called iCS-digital™ PSC. This assay combines a fast turnaround with high resolution, enabling the precise detection of sub-karyotypic defects such as the 20q11.21 chromosomal abnormality.
If you need to go further in genomic stability testing and detect structural arrangements such as balanced and unbalanced translocations, aneuploidy, inversions or duplications/deletions, we recommend our Duo iCS-Karyo. It is very comprehensive and remains affordable for most labs. If you require a more comprehensive approach including both SNV and CNV detection, the NGS-based Stem-Seq™ range is the one to turn to.
The Stem-Seq™ range includes the Stem-Seq™ Panel for SNV detection and the Stem-Seq™ Plus for SNV and CNV detection. This powerful technology will allow the detection of SNVs in 361 targeted locations and CNVs across the genome that may not have been previously considered or that could harbor unexpected variations. This comprehensive and unbiased view offers significant information on the genomic stability of your cells.
The ultimate combined solution for hPSC in-process genomic stability testing
Look deeper into any human cell. Detect both SNVs and CNVs
Cell line authentication
Mislabeling or cross-contamination creates risks of misidentification of cell lines. This happens more often than one might think: The International Cell Line Authentication Committee (ICLAC) keeps a registry of misidentified cell lines and to date, estimates that as many as 1/5 to 1/3 of known cell lines are wrongly identified. As of April 2024, their register of Misidentified Cell Lines holds records of 545 misidentified cell lines with no known authentic stock. It is therefore critical to check the cell line identity regularly throughout the time in culture. In terms of methodology, STR has been formally developed into an internationally recognized and accepted consensus standard for human cell line authentication and is the approach recommended by the ISSCR.
Need more information on cell authentication? We’re here to help.
Mycoplasma detection
Mycoplasma contamination can alter the capacity of pluripotent stem cells to differentiate properly and wreak havoc in labs. Unlike other bacteria, mycoplasma cannot be seen by observation only and can take months to get rid of.
It is therefore strongly recommended to regularly check for possible contamination and ensure the complete absence of mycoplasma in your cell culture.
Need more information on mycoplasma testing? We’re here to help.
Pluripotency & differentiation
Pluripotency is a key property of stem cells that allows them to differentiate into various cell types in the body. Molecular assays play a crucial role in studying pluripotency and differentiation stages by providing precise insights into gene expression profiles. This is why Stem Genomics has designed a new pluripotency assay that complements its range of services for scientists working on pluripotent stem cells.
Need more information on pluripotency and differentiation testing? We’re here to help.
One-stop-shop solution
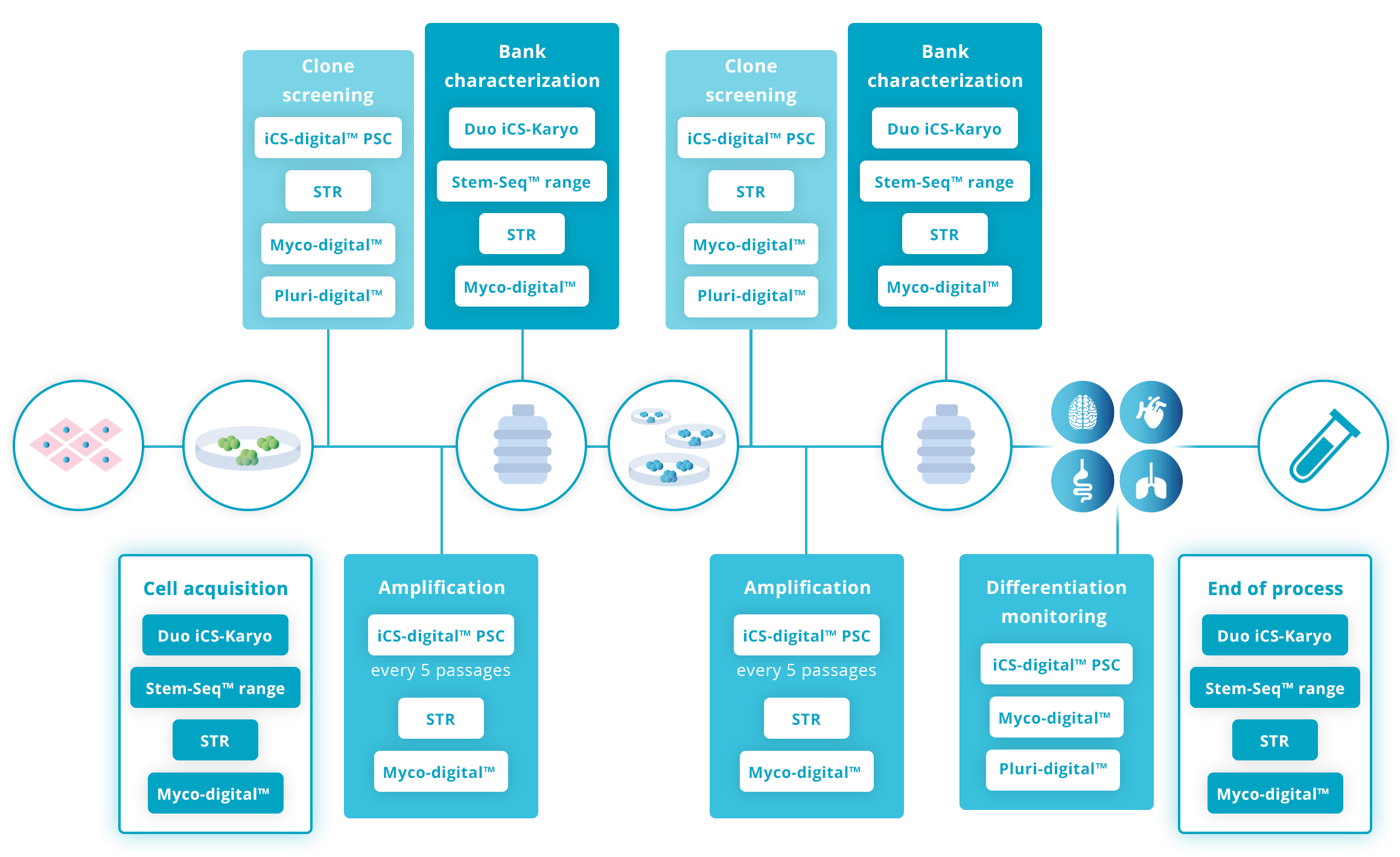
To increase your confidence in the quality of your cell line, Stem Genomics recommends regular genetic stability testing, mycoplasma detection and short tandem repeats throughout the lifecycle of the cells in culture. For other key stages, such as bank characterization, new cell line acquisition and end of process, we recommend adding G-Banding karyotyping to your QC. Finally, after clone screening and during differentiation, it is good practice to monitor pluripotency.
Watch a quick video presentation of the one-stop-shop service.
Streamline your stem cell research with a one-stop-shop solution for essential quality control
Need more information on one-stop-shop services? We’re here to help.
Recommended testing guidelines
The following guidelines are based on our extensive experience working with hPSC research scientists and recognized guidelines such as the ISSCR Standards for Human Stem Cell use in Research.
Definition
Pluripotent stem cells (PSCs). Cells capable of maintaining an undifferentiated state indefinitely and that give rise to all cells in the body.
Induced Pluripotent Stem Cells (iPSCs). A type of pluripotent stem cell derived from adult somatic cells and reprogrammed to an embryonic stem (ES) cell-like state by chemical or genetic reprogramming, commonly by inducing the expression of the transcription factors OCT4, SOX2, KLF4 and MYC (OSKM).
History
1981: Martin Evans of Cardiff University, UK, then at the University of Cambridge, is first to identify embryonic stem cells in mice.
1997: From the “Dolly the sheep” experiment, the first artificial animal clone, Iam Wilmut and his colleagues at the Roslin Institute, Edinburgh imagine that creating genetically matched tissue and organs in humans would be possible.
1998: James Thomson of the University of Wisconsin in Madison and John Gearhart of Johns Hopkins University in Baltimore isolate human embryonic stem cells and grow them in the lab.
2006: Shinya Yamanaka developed a revolutionary method for generating stem cells from existing cells of the body: inserting specific genes into the nuclei of adult cells, a process that results in the reversion of cells from an adult state to a pluripotent state.
2012: Nobel prize awarded to Shinya Yamanaka and John B. Gurdon for the discovery that mature cells could be reprogrammed.
This discovery marked a turning point in stem cell research, because it offered a way of obtaining human stem cells without the controversial use of human embryos.
Since then, iPSCs have demonstrated extraordinary applications in the fields of disease modelling, drug screening and regenerative medicine.
List of Stem Genomics' recommended assays for human Pluripotent Stem Cells:
- iCS-digital™ PSC range: available in 28 probes or 20q only, as a service or a kit, this unique test enables up to 93% detection of recurrent genomic abnormalities in hPSCs. More on iCS-digital™ PSC
- Duo iCS-Karyo: a powerful combination of traditional G-Banding karyotyping and iCS-digital™ PSC for comprehensive detection of PSC abnormalities. Available as a service only. More on Duo iCS-Karyo. G-banding alone also available. More on G-banding.
- The Stem-Seq™ range: a high-resolution, highly-sensitive, targeted NGS panel specifically designed for SNV and Indel detection combined with a pangenomic view of CNVs. This comprehensive test comes with various levels of reports and interpretations about the significant identified variants. More on the Stem-Seq™ range.
- Cell line authentication: easy, fast and affordable STR-based cell line authentication service in just 3 business days. More on the STR service.
- Pluri-digital™: a straightforward, affordable and fast solution that delivers accurate insight into gene expression profile. More on Pluri-digital™
- Mycoplasma detection: a robust detection method meeting the recommended regulatory guidance of 10 CFU/mL for 60 mycoplasma species. Results in just 3 days. More on Myco-digital.
- One-stop shop: perform up to 5 essential quality tests in days! Digital karyotyping, G-Banding, cell authentication, pluripotency and differentiation and mycoplasma testing in one-go! Benefit from bulk savings! More on the One-stop-shop service.

