The Stem-Seq™ range:
In-depth genomic knowledge
without the data overload
Expand your genomic knowledge using advanced sequencing technology
Human stem cells can develop molecular and genomic abnormalities, arising at a very low level of mosaicism. These defects, primarily Copy Number Variations (CNV) and Single Nucleotide Variants (SNV), need to be identified as quickly as possible in the process, as they can rapidly develop and overtake cell cultures. Their impact can be detrimental to the quality of the final product.
With the advent of cutting-edge technologies such as next-generation sequencing (NGS), the ability to detect novel abnormalities has greatly improved. Unlike traditional methods such as G-banded karyotyping, which are less sensitive and have limited resolution, NGS can detect variants with unprecedented precision. This new technology is making a significant contribution to our understanding of genomic variation and its potential impact in various clinical and research settings.
However, the main constraint of this method is the vast amount of information it can generate. This turns result interpretation into a time consuming and challenging exercise for most teams.
This is what led Stem Genomics to design a range of NGS-based sequencing assays with fully interpreted results that mean something to you.
Discover the Stem-Seq™ range in video
Watch short videos for a visual presentation of our latest NGS-targeted assays and discover the main benefits they offer scientists working on stem cells.
Stem-Seq™ Panel: Detect SNVs with a panel of 361 genes
Stem-Seq™ Plus: detect CNVs and SNVs in just one test.
A deeper understanding of targeted regions of interest
Based on innovative NGS technology, the Stem-Seq™ range enables scientists to target copy number variations (CNVs) throughout the genome and specific genes for single nucleotide variants (SNV detection). This is particularly suited to pluripotent stem cell scientists who encounter the highest level of mutations in their cells.
2 levels of tests:
- Stem-Seq™ Panel: a custom-made, targeted NGS panel composed of 361 genes that have been selected for their relevance to stem cell scientists. It detects Single-Nucleotide Variants (SNVs) associated with cancer (including TP53, BCOR, etc.) but also selected variants specific to pluripotent stem cells and their impact on the natural development of cells in culture.
- Stem-Seq™ Plus: it includes the Stem-Seq™ Panel for SNV detection as well as the additional capability to detect copy number variations (CNVs) across the entire genome. This powerful assay will allow the detection of CNVs in regions that may not have been considered previously or that could harbor unexpected variations. This comprehensive and unbiased view adds significant information on the genomic stability of your cells.
A clear and meaningful report
Stem-Seq™ Panel key specifications at a glance
- A wide range of 361 genes with known associations to cancer according to recognized database references (including TP53, etc.), and genes known to be recurrently mutated in hPSC (Salomonis N. et al. 2016, Merkle et al. 2020, Avior et al. 2020, Merkle et al. 2022).
- Deep sequencing of targeted genes with 1000x coverage, and a high resolution of 1% mosaicism/VAF to capture SNVs and Indels.
- Fully interpreted report.
- Delivery in 4-5 weeks (depending on the report option selected).
- Just send your gDNA sample at room temperature.
- Analysis available starting from 1 sample so you do not need to wait for other samples before processing begins!
All human cell types
Acquisition of a new line
Bank characterization
End of process
gDNA
Room temperature
1000 x
1%
4-5 weeks depending on report selection
Available from 1 sample
Stem-Seq™ Plus key specifications at a glance
- A comprehensive solution that combines both SNV and CNV detection in one assay.
- SNVs and Indels capture: deep sequencing of targeted genes with 1000x coverage, and a high resolution of 2% mosaicism/VAF.
- The panel contains 361 genes with known associations to cancer according to recognized database references (including TP53, etc.), and genes known to be recurrently mutated in hPSC (Salomonis N. et al. 2016, Merkle et al. 2020, Avior et al. 2020, Merkle et al. 2022).
- CNV detection: deep sequencing across the genome with 1000x and a high resolution of 15% mosaicism*.
- This sequencing approach uses a backbone and a Pool of Normal (PON) to enable a more accurate analysis of the large regions covered by CNVs and reduce false positives.
- Fully interpreted report.
- Delivery in 4 to 5 weeks, depending on the selected report.
- Just send your gDNA sample at room temperature.
- Analysis available starting from 1 sample so you do not need to wait for other samples before processing begins!
*A lower mosaicism can be obtained from the raw data that we will supply. However, we will not produce any interpretation under the 15-20% threshold.
All human cell types
Acquisition of a new line
Bank characterization
End of process
gDNA
Room temperature
1000 x
2% for SNVs
15% for CNVs*
300 kb
4-5 weeks depending on report selection
*A lower mosaicism can be obtained from the raw data that we will supply. However, we will not produce any interpretation under the 15% threshold.
Available from 1 sample
A comprehensive service to make your life easier
Stem Genomics handles the whole process for you from receipt of DNA samples to the delivery of the results. The library is built using Twist capture technology and the sequencing is performed on Illumina NovaSeq as paired-end reads. The final analysis will be sent to you within 4-5 weeks depending on the level of reporting you need or the type of analysis required (SNV only or SNV + CNV).
How it works:
Follow the step-by-step guidance to perform your assay: view useful documents
Practical testing guidelines
Recommended guidelines on genomic stability advise checking cells at various key steps in the workflow. Due to its exhaustivity and strong technical performance, the Stem-Seq™ range is best suited for the acquisition stage, when banking cells and/or at the end of the workflow. It strengthens the RUO process overall and supports the application for entering the clinical stage.
Click on the graphs below for more details
Guidelines for human Pluripotent Stem cell workflow
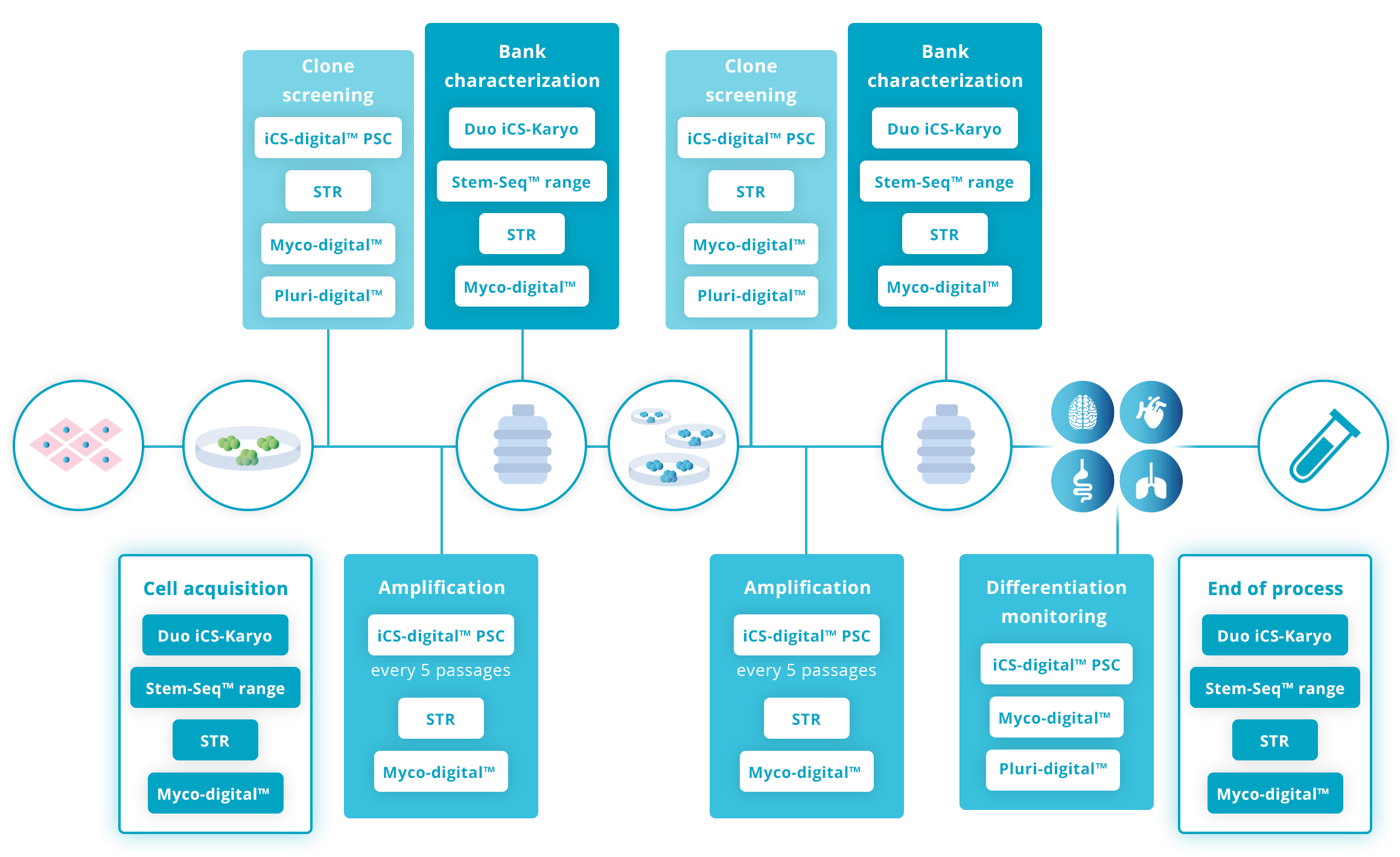
Guidelines for human Mesenchymal Stromal cell workflow
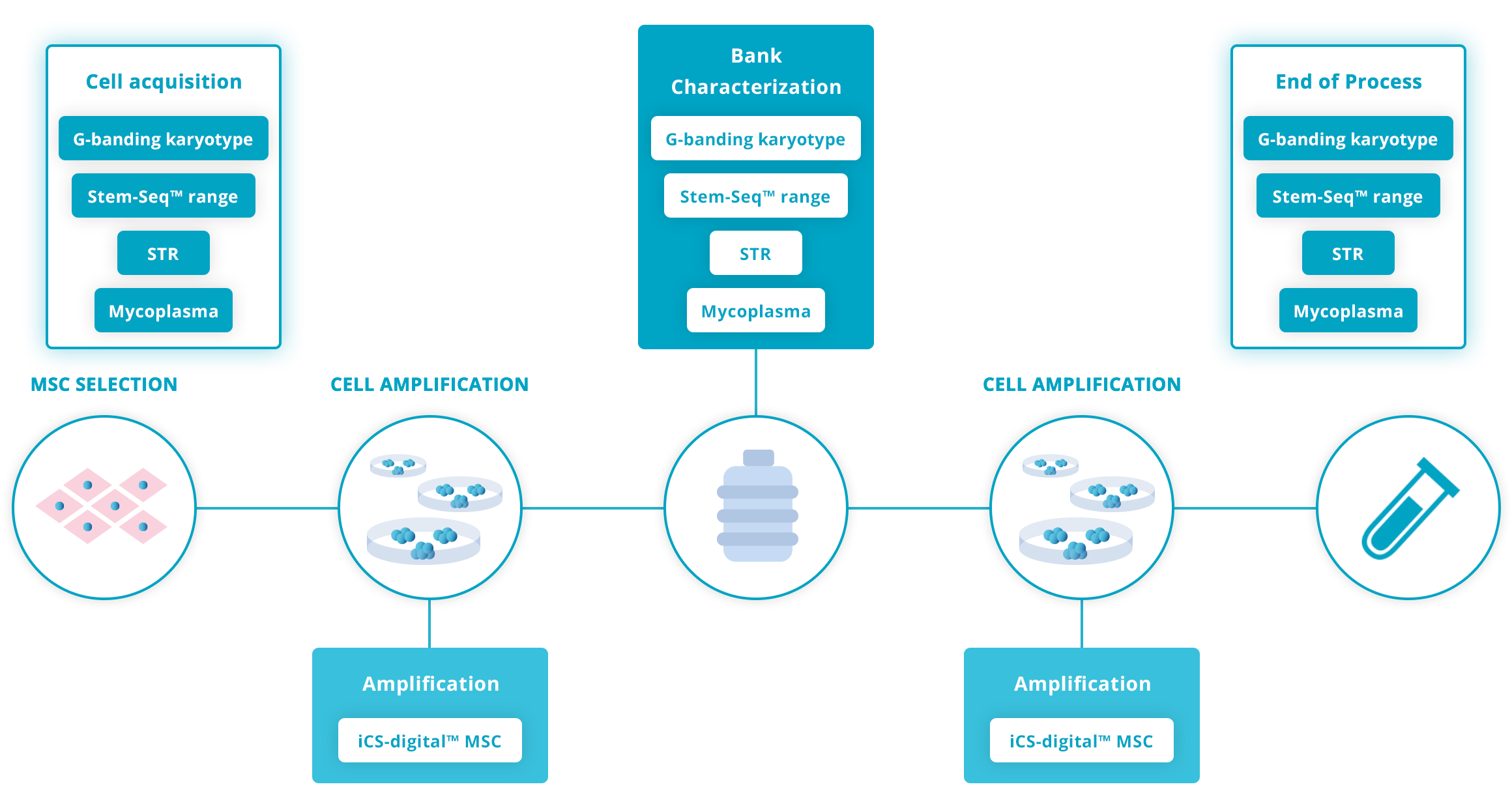
Amplification
Amplification

