Since their discovery in 2006 by Shinya Yamanaka, Induced Pluripotent Stem Cells (iPSCs) have demonstrated extraordinary applications in the fields of disease modelling, drugs screening and regenerative medicine. Increasingly popular with labs around the world, this promising field is nevertheless experiencing growing pains. « The biggest challenge we observe is the lack of rigorous quality controls throughout the cell culture process » says John De Vos, Professor at the Institute for Regenerative Medicine and Biotherapy (IRMB) and Director of the Cellular Therapy Department of the University Hospital Centre of Montpellier. In his work with Dr Said Assou in 2018 [1], 25 consecutive studies on iPSCs revealed that genomic testing was unsatisfactory, due to a lack of genomic integrity follow-up. More surprising is the speed in which novel genetic variant can become predominant in a culture, sometimes within 5 passages [2].
All facts supporting the need for regular check-ups.
Gap between the theory and practice: the challenges researchers face in the field
The gap between theory and practice in terms of regular quality controls seems to be linked to the labour and costs involved. However, those shortcuts could turn out to be false economies… with potential dramatic outcome.
Indeed, not routinely testing is a serious gamble. Chromosomic abnormalities are likely to happen at different steps during in vitro PSC culture: when reprogramming somatic cells, during cell culture or through genome editing (ref. Stem Genomics July 2021 article). In short, a simple “start and end workflow control” would not offer sufficient guarantees.
Pr. John De Vos says “It is always best to identify anomalies early in the process. If you do not test early enough, it is potentially impacting on the validity of your whole piece of research”. If not detected, acquired variants might interfere with studies that use iPSC cells for disease modelling or drug screening. At clinical levels, the risks can be more substantial if anomalies present in cells used for regenerative medicine, present variants involving genes such as TP53 or BCL2L1 commonly associated with cancer [3]. Other observed issues are: escaping apoptosis [4],[5], a decreased differentiation capacity [6], and faster cell cycling [7].
When a karyotype is not enough...
iPSCs are not genetically more unstable than somatic cells, however their high proliferation rate increases the risk of occurrence and then selection of genetic abnormalities. Therefore, they require close monitoring to ensure the integrity of the cell lines from beginning till the end process [2].
Some factors have already been reported to precipitate genomic change in PSCs and should be minimized: high density culture [8], enzymatic single-cell passaging and feederfree conditions [9].
Pr. John De Vos adds « When quality controls happen, we observe that most current assays such as karyotyping are not fully suitable for the required regular screening. The most recurrent genomic abnormality observed is not picked up by a karyotype analysis ».
Finding a practical solution
Taking into account that karyotyping can miss abnormalities smaller than 5-10 Mb, Prof John De Vos and Said Assou recognised the power of digital PCR (dPCR) in detecting the most frequent genomic anomaly, i.e. the 20q11.21 amplification [10], which accounts for 20% of PSC abnormalities detected worldwide. Although karyotyping is still important to detect balanced translocations and inversions, it is time consuming and therefore not suitable for in-process testing. Rapid tests for routine screening of cultured cells are mostly provided by PCR-based techniques [11]. Using specific primer sets and fluorescent probes, dPCR amplifies targeted genomic fragments in thousands of individual reactions, thus allowing nucleic acid quantification and detection with high precision. This is making dPCR very attractive to genetics researchers (ref. Stem Genomics April 2021 article).
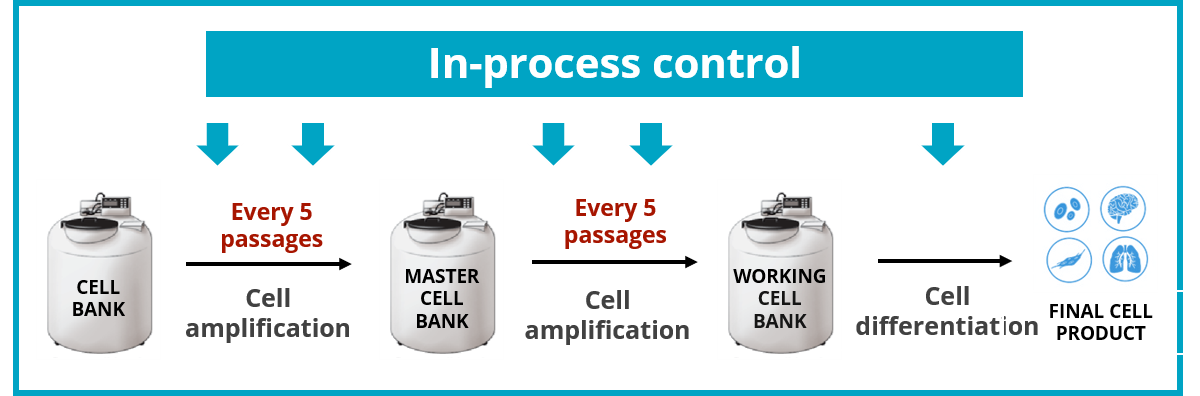
Good practice would be to have an initial characterization including karyotyping and dPCR, and, for more critical investigations, also include a microarray assay and TP53 gene sequencing. Routine surveillance is then mandatory at a frequency that is important to maintain close to 5 passages, which is only made possible by using the dPCR technology.
All in all, early detection of abnormalities clearly prevents the investment of time and money in maintaining abnormal iPSC cultures. Beyond the financial aspects, this also ensures that the iPSCs remain the faithful copy of the cells from which they derive, lowering the risks of working on a transformed model and securing your final cell product.
[1] Assou, S., Bouckenheimer, J., and De Vos, J. (2018). Concise review: assessing the genome integrity of human induced pluripotent stem cells: what quality control metrics? Stem Cells 36, 814–821.
[2] Draper JS, Smith K, Gokhale P et al. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat. Biotechnol. 22, 53–54 (2004)
[3] Andrew, P.W. (2021) Human pluripotent stem cells: genetic instability or stability? Regenerative Medecine,16 (2).Pp.113-115.
[4] Avery S, Hirst AJ, Baker D, et al. BCL-XL mediates the strong selective advantage of a 20q11.21 amplification commonly found in human embryonic stem cell cultures. Stem Cell Reports. 2013;1(5):379-386. Published 2013 Oct 31.
[5] Merkle FT, Ghosh S, Kamitaki N, et al. Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations. Nature. 2017;545(7653):229-233.
[6] Markouli C, Couvreu De Deckersberg E, Regin M, et al. Gain of 20q11.21 in Human Pluripotent Stem Cells Impairs TGF-βDependent Neuroectodermal Commitment. Stem Cell Reports. 2019;13(1):163-176.
[7] Barbaric I, Biga V, Gokhale PJ, et al. Time-lapse analysis of human embryonic stem cells reveals multiple bottlenecks restricting colony formation and their relief upon culture adaptation. Stem Cell Reports. 2014;3(1):142-155. Published 2014
[8] Jacobs K, Zambelli F, Mertzanidou A, et al. Higher-Density Culture in Human Embryonic Stem Cells Results in DNA Damage and Genome Instability. Stem Cell Reports. 2016;6(3):330-341.
[9] Garitaonandia I., Amir H., Sesillo Boscolo F. , Wambua G. H.,Schultheisz H. L., Sabatini K., Morey R., Waltz, Wang Y. C., Tran H., Leonardo T. R., Nazor K., Slavin I., Lynch C., Li Y., Coleman R., Gallego Romero I., Altun G., Reynolds D., Dalton S.,
Parast M., Loring F. J. ,Laurent L. C. (2015). Increased Risk of Genetic and Epigenetic Instability in Human Embryonic Stem Cells Associated with Specific Culture Conditions, Plos one.
[10] Assou S., Girault N., Plinet M., Bouckenheimer J., Sansac C., Combe M., Mianne J., Bourguignon C., Fieldes M., Ahmed E., Commes T., Boureux A., Lemaître JM., and De Vos J. (2020) Stem Cell Reports, Vol. 14 , 1-8.
[11] Baker D., Hirst A. J., Gokhale P. J., Juarez M. A., Williams S., Wheeler M., Bean K., Allison T. S., Moore H. D., Andrews P.W., Barbaric I. Detecting Genetic Mosaicism in Cultures of Human Pluripotent Stem Cells (2016) Stem Cell Reports.