One of the downsides of the amazing abilities pluripotent stem cells have to multiply indefinitely is their propensity at developing genomic defects during their time in culture. For this reason, any scientist working with human pluripotent stem cells (hPSC) is faced with a challenge that is to find the most suitable testing strategy for their needs. In this article, we highlight the most important chromosomal anomalies you should watch for and share our expertise in this complex field of integrity testing.
Chromosome 20q11.21. the most common genetic defect in hPSCs
Back in 2011, the International Stem Cell Initiative analysed 125 human embryonic stem (ES) cell lines and 11 induced pluripotent stem (iPS) cell lines, from 38 laboratories worldwide looking for genetic changes over the duration of the cells’ culture.
Most lines were analysed at early and late passages revealing that there was a progressive tendency for genomic defects to develop later down the line, commonly affecting chromosomes 1, 12, 17 and 20 (Amps et al., 2011). The most regular occurrence is a copy number gain on chromosome 20q11.21 from which BCL2L1, an inhibitor of apoptosis, has been identified as the driver gene (Avery et al., 2013) Abnormal overexpression of the antiapoptotic gene BCL2L1 confers a survival advantage to the cells and is often commonly observed in human cancer (Beroukhim et al., 2010; Scotto et al., 2008; Tabach et al., 2011)
More specifically, chromosome 20q amplification is strongly correlated with colorectal adenoma-to-carcinoma progression, and BCL2L1 may have a functional role in this transition (Sillars-Hardebol et al., 2012) . In addition, there is also an inverse relationship between BCL2L1 expression and the chemosensitivity of cancer cell lines (Amundson et al., 2000). This contributes to the cells’ chemoresistance (Williams et al., 2005).
Gain of the chromosome 20q also leads to a decreased differentiation potential toward neural lineages (Werbowetski-Ogilvie et al., 2009)
Therefore, it is essential to identify the potential presence of 20q anomaly to eradicate the detrimental impact at clinical level but also for basic research, disease modelling and pharmaceutical efficacy and toxicity assays.
At Stem Genomics, we have found very similar trends with the cell lines we have tested so far. Looking at the samples analysed, 26% showed a recurrent abnormality, with 50% of the abnormalities found on the 20q chromosome. Elena Hauser, Platform Manager adds: “The most frequent recurrent abnormality found in the PSC lines tested by our platform has consistently been the 20q since we started testing back in 2018”.
As for the 20q11.21 region, other recurrent abnormalities encountered in PSCs are often associated with risk of cancer. Functional consequences of those recurrent abnormalities are the ability to escape apoptosis (Avery et al., 2013), decrease differentiation capacity (Markouli et al., 2019) increase tumorigenicity (Ben-David, 2015) and faster cell-cycling (Barbaric et al., 2014).
This is where research stands today, however, with the continuous improvement of the PSC characterisation methods as well as the adoption of new culture conditions, it is to be expected that new abnormalities will get identified. This means that tests to detect these abnormalities and their interpretation should be regularly updated to match ground reality.
Identifying the most suitable test combination
The risk of genomic alteration increases with the time PSCs are maintained in culture. In their 2011 study, the International Stem Cell Initiative observed that 77% of the 20q gains were happening in late passages and only 23% in early passages. Additionally, none of the abnormalities found in early passages disappeared over time, reflecting the selective advantage of the acquired abnormalities. This supports the need for regular testing throughout the culture process.
The challenge for researchers is to find the right test strategy in the multitude of technologies available, but also the right frequency, sometimes a fine balance between risks, time and costs.
To help you establish the best solution to meet your needs, we’ve asked the Technical team here at Stem Genomics what they advised.
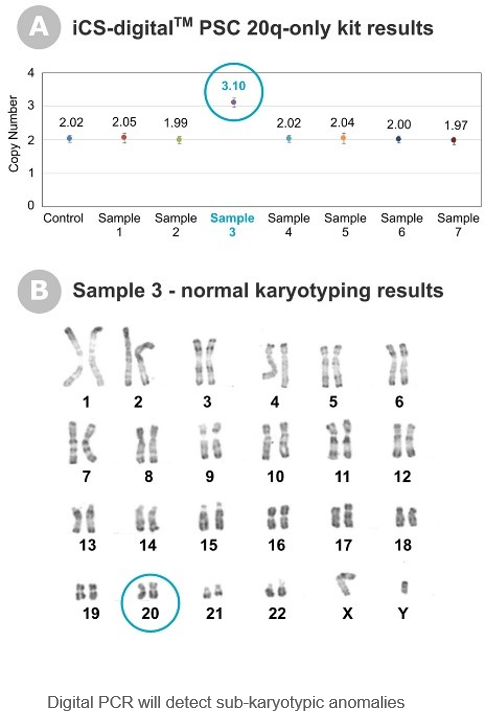
“The most frequent technique combination our clients are using is the association of a karyotype looking at the whole genome in combination with our targeted digital PCR test to identify specific PSCs’ anomalies below 5Mb that the karyotype cannot detect. The FISH technology is another possible targeted approach but it can be laborious with concerns over tandem duplications detections (Mascarello et al., 2011)
Considering the speed a recurrent abnormality is able to take over the culture, we and others recommend (and this is also found in (McIntire et al., 2020) to test at least every 5-10 passages.
Although a Karyotype is the right approach for cell line characterisation at early, late or banking stage, it usually takes too long and is therefore not suitable for in-process testing.
“The need for frequent testing combined with a high-resolution detection capacity make the digital PCR technology an excellent technical choice. The technology amplifies targeted genomic fragments in thousands of individual reactions, allowing nucleic acid quantification and detection with high precision. In addition, the technology is fast (1 to 3 days) and reasonable in costs compared with other technologies. All in all it’s perfectly adapted to the requirements of regular testing on PSC cell lines which is why we’ve fully embraced this technology to meet our clients’ needs “, says Elena Hauser, Platform Manager at Stem Genomics.